Integrative structural biology
Team 4 – UMR1167 – Inserm – Lille University – CHU Lille – Institut Pasteur de Lille
Presentation
Our team investigates structural basis and molecular mechanisms underlying protein biological functions, and the impact of their deregulation in human diseases.
In particular, we are interested in intrinsically disordered proteins. These proteins exhibit specific characteristics such as numerous post-translational modifications and regions rich in proline residues, modulating their interactions, as well as their propensity to aggregate in proteinopathies.
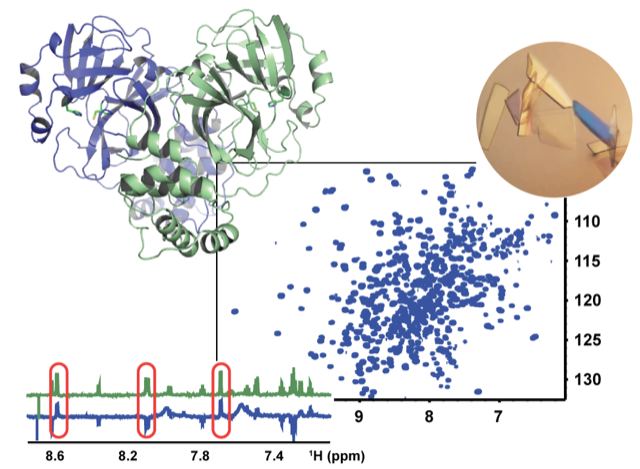
At the heart of our work are biophysical multidisciplinary methods for characterizing the structure of proteins, as well as the interactions between them, such as Nuclear Magnetic Resonance spectroscopy, X-ray cristallography and cryo-electron microscopy.
Our research is essentially fundamental, but we remain attentive to its application to the development of new therapeutic approaches, the evaluation of the therapeutic potential of various compounds from small molecules to nanobodies (very small antibodies), and the study of the mode of action of bioactive molecules.
Highlights
- 2020 ANR URANUS Project JCJC, Davy Sinnaeve.
Understanding the function of flexible biopolymers such as disordered proteins, glycans or oligonucleotides requires insight in their conformational ensembles. Liquid-state NMR is the leading technique reporting on this. Unfortunately, fast structural averaging on the NMR time scale strongly reduces chemical shift dispersion, which, given the many 1H-1H couplings, results in spectral overlap. This obstructs resolving 1H NMR data, such as residual dipolar couplings (RDCs), that report on long-range structural order and allosteric changes. In addition, molecular flexibility has until now greatly complicated the interpretation of RDC data. URANUS proposes a general strategy to solve both these issues for any biopolymer. On the mid-term, we will create a widely accessible toolbox of techniques that is generally applicable to any biopolymer.
- 2019 ANR CREST Project JCJC, Robert Schneider,
Voltage-gated ion channels (VGICs) mediate electrical signaling in cells. The resting state of their voltage-sensitive domains (VSDs) remains unknown, since it is only adopted in the presence of the membrane’s resting potential. CREST proposes to address this problem with an innovative approach: establishing a Donnan potential across the membrane of liposomes via the asymmetric presence of impermeable polyelectrolytes and studying VGICs reconstituted in these liposomes by solid-state NMR.
- The Distalz Labex has been renewed for the period 2020 to 2025, in a second round of the project in which the team will participate. One of the major achievements of DISTALZ is the complementarity of its eight teams : geneticists are in contact with molecular and cellular biologists, and all of them are connected with biophysicists, clinicians, psychologists and ethicists who work together on a daily basis. This makes it possible to join forces in a single laboratory with a common research program, in order to overcome obstacles of AD research, accelerate discovery of innovative solutions based on new potential drug targets, and translate these results into therapeutic strategies that are socially and ethically viable.
Transversal project
 Xavier Hanoulle has participated in the interdisciplinary efforts of the Pasteur Institute campus against COVID-19, and has obtained support from the Isite for his project 3CLPro.
Xavier Hanoulle has participated in the interdisciplinary efforts of the Pasteur Institute campus against COVID-19, and has obtained support from the Isite for his project 3CLPro.
In the context of the SARS-CoV-2 pandemic, this project aims to rapidly identify molecules or molecule fragments that could inhibit the 3CLpro protease of SARS-CoV-2. The 3CLpro protease is a non-structural protein of the coronavirus which is essential in its replication cycle. Its enzymatic proteolytic activity matures the polyproteins pp1a and pp1b. The mature proteins then constitute the replication complexes of the virus. 3CLpro therefore represents a therapeutic target. Molecules capable of inhibiting 3CLpro can either bind to its active site or to its dimerization interface, since this enzyme is active as a dimer. In this project, we propose to perform a molecule / fragment screening on the 3CLpro protease of SARS-CoV-2 by nuclear magnetic resonance (NMR) spectroscopy. The realization of this project is based on fragment libraries available within the U1177 research unit (B. Desprez, Institut Pasteur de Lille). The molecules identified in the project will first be tested in an in vitro activity test, then in a cellular system (J. Dubuisson, Center for Infection and Immunity of Lille, Institut Pasteur de Lille), in order to validate their potential efficacy against SARS-CoV-2.
Members
Isabelle LANDRIEU
DR2 CNRS, group leader
Marc AUMERCIER
DR2 CNRS
Laurent COUTTE
MCU Univ Paris XI
Xavier HANOULLE
DR2 CNRS
Didier MONTE
CRHC CNRS
Robert SCHNEIDER
MCU Univ Lille
Davy SINNAEVE
CRCN CNRS
Caroline SMET-NOCCA
MCU Univ Lille, CNRS
Alexis VERGER
CRCN CNRS
Vincent VILLERET
DR1 CNRS
François-Xavier CANTRELLE
IGN UnivLille
Bernard CLANTIN
IR1 CNRS
Frédérique DEWITTE
IECN CNRS
Zoé LENS
IECN CNRS
Justine MORTELECQUE
IE CNRS, CDD
Guillaume ROUAULT
IE CNRS, CDD
Emmanuelle BOLL
TCS CNRS
Elian DUPRE
Post-doc UnivLille
Orgeta ZEJNELI
PhD student, cotutelle Luc Buée
Danai MOSCHIDI
PhD student
Publications
Monté D, Clantin B, Dewitte F, Lens Z, Rucktooa P, Pardon E, Steyaert J, Verger A, Villeret V.
Crystal structure of human Mediator subunit MED23.
Nat Commun. 2018 Aug 23;9(1):3389. doi: 10.1038/s41467-018-05967-y.
Dujardin M, Madan V, Gandhi NS, Cantrelle FX, Launay H, Huvent I, Bartenschlager R, Lippens G, Hanoulle X.
Cyclophilin A allows the allosteric regulation of a structural motif in the disordered domain 2 of NS5A and thereby fine-tunes HCV RNA replication.
J Biol Chem. 2019 Aug 30;294(35):13171-13185. doi: 10.1074/jbc.RA119.009537.
Sinnaeve D, Ilgen J, Di Pietro ME, Primozic JJ, Schmidts V, Thiele CM, Luy B.
Probing Long-Range Anisotropic Interactions: a General and Sign-Sensitive Strategy to Measure 1 H-1 H Residual Dipolar Couplings as a Key Advance for Organic Structure Determination.
Angew Chem Int Ed Engl. 2020 Mar 23;59(13):5316-5320. doi: 10.1002/anie.201915278.
Despres C, Di J, Cantrelle FX, Li Z, Huvent I, Chambraud B, Zhao J, Chen J, Chen S, Lippens G, Zhang F, Linhardt R, Wang C, Klärner FG, Schrader T, Landrieu I, Bitan G, Smet-Nocca C.
Major Differences between the Self-Assembly and Seeding Behavior of Heparin-Induced and in Vitro Phosphorylated Tau and Their Modulation by Potential Inhibitors.
ACS Chem Biol. 2019 Jun 21;14(6):1363-1379. doi: 10.1021/acschembio.9b00325.
Wolter M, de Vink P, Neves JF, Srdanović S, Higuchi Y, Kato N, Wilson A, Landrieu I, Brunsveld L, Ottmann C.
Selectivity via Cooperativity : Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J Am Chem Soc. 2020 Jul 8;142(27):11772-11783. doi: 10.1021/jacs.0c02151.
Keywords
Protein structure ; Protein-Protein interactions ; intrinsically disordered proteins ; Protein post-translational modifications ; Ligand-Protein interactions ; Nuclear Magnetic Resonance Spectroscopy ; Cristallography ; Transdisciplinary research
Team contact
Isabelle Landrieu
CNRS Research director
isabelle.landrieu@univ-lille.fr
03 20 87 73 02 ; 03 62 53 17 12